This page is part of the Observations of notifiable communicable infectious diseases (v1.13.1: Release) based on FHIR (HL7® FHIR® Standard) R4 . This is the current published version. For a full list of available versions, see the Directory of published versions
Home
Official URL : http://fhir.ch/ig/ch-elm/ImplementationGuide/ch.fhir.ig.ch-elm
Version :
1.13.1
Active
as of 2026-01-18
Computable Name : CH_ELM
Copyright/Legal : CC0-1.0
Introduction
CH ELM is a project of the Swiss Federal Office of Public Health (FOPH), Communicable Diseases Division, to enable laboratories to send their observations of notifiable communicable infectious diseases to the FOPH electronically.
A report is created as a specialized Clinical Document based on the HL7® FHIR® standard. This FHIR document is sent to the FOPH by a FHIR RESTful web API endpoint . CH ELM derives from the Swiss implementation guides and the European laboratory project (see graphical overview ).
The expected content of the FHIR document, based on the ordinance of the Federal Office of Public Health (DE , FR , IT ), is defined in the logical model . A mapping shows how to access the data from the FHIR document. In addition, further documentation for specific topics can be found on the guidance page and the use cases describe the different scenarios with respective examples for specific organisms.
The specification herewith documented is work in progress. No liability can be inferred from the use or misuse of this specification, or its consequences.
Changelog with significant changes, open and closed issues.
Download : You can download this implementation guide in npm format from here .
Implementation Support for Laboratories
This guide supports you as a laboratory in the following way:
Use cases describe the different scenarios with respective example reports for specific organisms, e.g. for Neisseria gonorrhoeae in xml or json , you choose if you want to provide the FHIR laboratory report in xml or json format.What needs to be defined in the laboratory report is defined in FHIR document , profiles define the constraints on FHIR resources which need to be included in the report, see the overview here .
The data elements defined by the ordinance of the Federal Office of Public Health (FOPH) for the report are defined in the logical model and are then mapped to the FHIR document structure.Value sets for the terminology are provided as FHIR resources which you can directly import from the npm package .
The laboratory report is sent to the FOPH by a FHIR RESTful web API endpoint .
Further documentation for specific topics can be found on guidance and/or by contacting the FOPH directly.
FHIR R4 has a huge implementation community and offers various libraries to support the implementation of FHIR based solutions, for creating the FHIR document or providing a client for doing the FHIR API calls. For java we recommend hapi-fhir , for .NET firely-net-sdk , but there are also multiple other options . If you have questions about general FHIR questions do not hesitate to ask in chat.fhir.org .
To check if your report is valid to the requirements of this implementation guide you can check it with the FHIR Validator providing this package as a parameter and specifying the profile http://fhir.ch/ig/ch-elm/StructureDefinition/ch-elm-document-strict.
Since the FHIR API is standardized you can test your client also against a public FHIR test server like hapi .
If you have general feedback this implementation guide you find at the bottom a "Propose a change" link where you can raise an issue.
Must Support
For the CH ELM exchange format, the mustSupport flag set to true has the following meaning:
IP Statements
HL7®, HEALTH LEVEL SEVEN®, FHIR® and the FHIR
This publication includes IP covered under the following statements.
CC0-1.0 Show Usage
ISO maintains the copyright on the country codes, and controls its use carefully. For further details see the ISO 3166 web page: https://www.iso.org/iso-3166-country-codes.html Show Usage
ISO 3166-1 Codes for the representation of names of countries and their subdivisions — Part 1: Country code : BfsCountryCodes , Bundle/10Doc-Legionella ... Show 217 more , Bundle/11Doc-Malaria , Bundle/12Doc-Mpox , Bundle/13Doc-Shigella , Bundle/14Doc-Neisseriameningitidis-confirmationtest , Bundle/14Doc-Neisseriameningitidis-confirmationtest-originalorder , Bundle/15Doc-Measles-Seroconversion , Bundle/16Doc-Dengue-Titer , Bundle/17Doc-Neisseria , Bundle/18Doc-C-diphtheriae , Bundle/19Doc-S-pneumoniae , Bundle/1Doc-NeisseriaGonorrhoeae , Bundle/1bDoc-NeisseriaGonorrhoeae , Bundle/1cDoc-NeisseriaGonorrhoeae , Bundle/20Doc-Vibrio-cholerae , Bundle/21Doc-HepatiteE , Bundle/22Doc-H-influenzae , Bundle/23Doc-F-tularensis , Bundle/24Doc-Chikungunya , Bundle/25Doc-Tick-borne-encephalitis , Bundle/26Doc-HepatiteA , Bundle/28Doc-Listeria-monocytogenes , Bundle/29Doc-Rubella , Bundle/2Doc-ChlamydiaTrachomatis , Bundle/2Doc-ChlamydiaTrachomatis-Vct , Bundle/30Doc-Salmonella-enteritidis , Bundle/32Doc-Rubella-avidity , Bundle/33Doc-Salmonella-valueString , Bundle/34Doc-Brucella , Bundle/35Doc-CJD , Bundle/36Doc-Salmonella-paratyphi , Bundle/37Doc-Zika , Bundle/38Doc-Anthrax , Bundle/39Doc-Botulims , Bundle/3Doc-CPE , Bundle/40Doc-Crimean-Congo , Bundle/41Doc-Ebola , Bundle/42Doc-Lassa , Bundle/43Doc-Marburg , Bundle/44Doc-Mers-CoV , Bundle/45Doc-Sars-CoV , Bundle/46Doc-Yersinia-pestis , Bundle/47Doc-Variola , Bundle/48Doc-Mpox-Clade , Bundle/49Doc-HIV , Bundle/4Doc-Campylobacter , Bundle/50Doc-HIV-viremia , Bundle/51Doc-Gelbfieber , Bundle/52Doc-Hantavirus , Bundle/53Doc-InfluenzaHxNy , Bundle/54Doc-Poliomyelitis , Bundle/55Doc-Tollwut , Bundle/56Doc-Trichinella-spiralis , Bundle/57Doc-West-Nile , Bundle/58Doc-Coxiella , Bundle/59Doc-EHEC , Bundle/5Doc-TreponemaPallidum , Bundle/61Doc-Tb-Susceptibility , Bundle/63Doc-Tb-Genotyping , Bundle/65Doc-Tuberculosis , Bundle/66Doc-HivRecency , Bundle/67Doc-Emerging-Pathogen , Bundle/6Doc-Influenza , Bundle/7Doc-SARSCoV2 , Bundle/8Doc-HepatiteB , Bundle/9Doc-HepatiteC , CHElmHumanName , CH_ELM , CH_ELM_CapabilityStatement_DocumentRecipient , ChElmCodeableConcept , ChElmCoding , ChElmComposition , ChElmCompositionStrict , ChElmDiagnosticReport , ChElmDocument , ChElmDocumentStrict , ChElmExpectingOrganismSpecification , ChElmExpectingOrganismSpecificationToResultsCompletionVs , ChElmExpectingSpecimenSpecification , ChElmExpectingSpecimenSpecificationToResultsCompletionVs , ChElmExtDepartment , ChElmExtHivCode , ChElmExtVctCode , ChElmFophBusinessRules , ChElmFophCodeReserve , ChElmFophPatientNameRepresentation , ChElmInterpretationCodesAvidity , ChElmInterpretationCodesPos , ChElmInterpretationCodesPosNeg , ChElmInterpretationCodesResSus , ChElmInterpretationCodesSero , ChElmInterpretationCodesTiter , ChElmInterpretationCodesVs , ChElmLabStudyTypes , ChElmLaboratoryReport , ChElmObservationComponentConcentration , ChElmObservationProfileVs , ChElmObservationResultsLaboratory , ChElmObservationResultsLaboratoryGenotyping , ChElmObservationResultsLaboratoryGenotypingStrict , ChElmObservationResultsLaboratoryHivRecencyStrict , ChElmObservationResultsLaboratoryMicrobiology , ChElmObservationResultsLaboratoryMicrobiologyStrict , ChElmObservationResultsLaboratorySusceptibility , ChElmObservationResultsLaboratorySusceptibilityStrict , ChElmObservationRto , ChElmObservationTxt , ChElmObservationVirl , ChElmOrganizationAuthor , ChElmOrganizationLab , ChElmOrganizationOrderer , ChElmPatient , ChElmPatientHIV , ChElmPatientInitials , ChElmPatientVCT , ChElmPractitionerOrderer , ChElmPractitionerRoleOrderer , ChElmQuantityArbU , ChElmResultsBotOrg , ChElmResultsBotSpec , ChElmResultsBruOrg , ChElmResultsCampDiarOrg , ChElmResultsCampOrg , ChElmResultsCholOrg , ChElmResultsCjdOrg , ChElmResultsCodedValuesLaboratory , ChElmResultsCompleteSpec , ChElmResultsCompletionVs , ChElmResultsComponentAntibioticCpe , ChElmResultsComponentAntibioticTb , ChElmResultsComponentGeneCpe , ChElmResultsComponentGeneTb , ChElmResultsComponentHivRecency , ChElmResultsComponentVs , ChElmResultsDiphOrg , ChElmResultsDiphSpec , ChElmResultsEbolOrg , ChElmResultsEhecOrg , ChElmResultsEhecToxOrg , ChElmResultsGeniSpec , ChElmResultsGenoToComponentCode , ChElmResultsGenoToComponentInterpretationCode , ChElmResultsHaemOrg , ChElmResultsHantaOrg , ChElmResultsHivOrg , ChElmResultsInfOrg , ChElmResultsInfluenzaHxnyOrg , ChElmResultsLaboratoryObservation , ChElmResultsLaboratoryObservationComplete , ChElmResultsLaboratoryObservationGeno , ChElmResultsLaboratoryObservationSusc , ChElmResultsLegOrg , ChElmResultsLisOrg , ChElmResultsLisSpec , ChElmResultsMalOrg , ChElmResultsMaldiTofOrg , ChElmResultsMeaOrg , ChElmResultsMenOrg , ChElmResultsMpoxOrg , ChElmResultsMpxCtng , ChElmResultsMpxSash , ChElmResultsMpxSashec , ChElmResultsPneuOrg , ChElmResultsSalOrg , ChElmResultsSalOrgComplete , ChElmResultsShiOrg , ChElmResultsSterileSpec , ChElmResultsSuscToComponentCode , ChElmResultsSuscToComponentInterpretationCode , ChElmResultsSuscToComponentObservationProfile , ChElmResultsToFophPatientNameRepresentation , ChElmResultsToInterpretationCode , ChElmResultsToObservationProfile , ChElmResultsTubGenOrg , ChElmResultsTubOrg , ChElmResultsTubSpec , ChElmResultsTulOrg , ChElmResultsVirusCultOrg , ChElmServicePreviousRequestLaboratoryOrder , ChElmServiceRequestLaboratoryOrder , ChElmSpecimen , ChElmStatus , ChExtElmStatus , DocumentReference/1-DocumentReferenceStrict , DocumentReference/1c-DocumentReferenceStrict , DocumentReference/2-DocumentReference , DocumentReference/2-DocumentReferenceStrict , DocumentReference/2-DocumentReferenceVctStrict , DocumentReference/Publish-1Doc-NeisseriaGonorrhoeae , DocumentReference/Publish-2Doc-ChlamydiaTrachomatis-Vct , DocumentReference/Publish-4Doc-Campylobacter , DocumentReference/Publish-5Doc-TreponemaPallidum , DocumentReference/Publish-6Doc-Influenza , DocumentReference/Publish-7Doc-SARSCoV2 , FindDocumentReferencesResponse , IDN , IdnIdentifier , Patient/Pat-001 , Patient/Pat-003 , Patient/Pat-004 , Patient/Pat-005 , Patient/Pat-006 , Patient/Pat-007 , Patient/Pat-VCT , PublishDocumentReference , PublishDocumentReferenceResponse , PublishDocumentReferenceStrict , Test100Kosovo , Test101InvalidCountryCode , Test92WarningBerUidGln , Test93ErrorFullNameInfluenca , Test94IgnoreSourceWarning , Test95IgnoreObservationInterpretationCodesWarning , Test96IgnoreObservationInterpretationCodesInformation , Test97CoaxiellaNoRatio , Test98HivInvalidCode , Test99CoaxiellaZeroDenominator and elmstatus
The UCUM codes, UCUM table (regardless of format), and UCUM Specification are copyright 1999-2009, Regenstrief Institute, Inc. and the Unified Codes for Units of Measures (UCUM) Organization. All rights reserved. https://ucum.org/trac/wiki/TermsOfUse Show Usage
This material contains content from LOINC . LOINC is copyright © 1995-2020, Regenstrief Institute, Inc. and the Logical Observation Identifiers Names and Codes (LOINC) Committee and is available at no cost under the license . LOINC® is a registered United States trademark of Regenstrief Institute, Inc. Show Usage
LOINC : Bundle/10Doc-Legionella , Bundle/11Doc-Malaria ... Show 105 more , Bundle/12Doc-Mpox , Bundle/13Doc-Shigella , Bundle/14Doc-Neisseriameningitidis-confirmationtest , Bundle/14Doc-Neisseriameningitidis-confirmationtest-originalorder , Bundle/15Doc-Measles-Seroconversion , Bundle/16Doc-Dengue-Titer , Bundle/17Doc-Neisseria , Bundle/18Doc-C-diphtheriae , Bundle/19Doc-S-pneumoniae , Bundle/1Doc-NeisseriaGonorrhoeae , Bundle/1bDoc-NeisseriaGonorrhoeae , Bundle/1cDoc-NeisseriaGonorrhoeae , Bundle/20Doc-Vibrio-cholerae , Bundle/21Doc-HepatiteE , Bundle/22Doc-H-influenzae , Bundle/23Doc-F-tularensis , Bundle/24Doc-Chikungunya , Bundle/25Doc-Tick-borne-encephalitis , Bundle/26Doc-HepatiteA , Bundle/28Doc-Listeria-monocytogenes , Bundle/29Doc-Rubella , Bundle/2Doc-ChlamydiaTrachomatis , Bundle/2Doc-ChlamydiaTrachomatis-Vct , Bundle/30Doc-Salmonella-enteritidis , Bundle/32Doc-Rubella-avidity , Bundle/33Doc-Salmonella-valueString , Bundle/34Doc-Brucella , Bundle/35Doc-CJD , Bundle/36Doc-Salmonella-paratyphi , Bundle/37Doc-Zika , Bundle/38Doc-Anthrax , Bundle/39Doc-Botulims , Bundle/3Doc-CPE , Bundle/40Doc-Crimean-Congo , Bundle/41Doc-Ebola , Bundle/42Doc-Lassa , Bundle/43Doc-Marburg , Bundle/44Doc-Mers-CoV , Bundle/45Doc-Sars-CoV , Bundle/46Doc-Yersinia-pestis , Bundle/47Doc-Variola , Bundle/48Doc-Mpox-Clade , Bundle/49Doc-HIV , Bundle/4Doc-Campylobacter , Bundle/50Doc-HIV-viremia , Bundle/51Doc-Gelbfieber , Bundle/52Doc-Hantavirus , Bundle/53Doc-InfluenzaHxNy , Bundle/54Doc-Poliomyelitis , Bundle/55Doc-Tollwut , Bundle/56Doc-Trichinella-spiralis , Bundle/57Doc-West-Nile , Bundle/58Doc-Coxiella , Bundle/59Doc-EHEC , Bundle/5Doc-TreponemaPallidum , Bundle/61Doc-Tb-Susceptibility , Bundle/63Doc-Tb-Genotyping , Bundle/65Doc-Tuberculosis , Bundle/66Doc-HivRecency , Bundle/67Doc-Emerging-Pathogen , Bundle/6Doc-Influenza , Bundle/7Doc-SARSCoV2 , Bundle/8Doc-HepatiteB , Bundle/9Doc-HepatiteC , ChElmComposition , ChElmCompositionStrict , ChElmExpectingOrganismSpecification , ChElmExpectingSpecimenSpecification , ChElmLabStudyTypes , ChElmObservationResultsLaboratory , ChElmObservationResultsLaboratoryGenotyping , ChElmObservationResultsLaboratoryGenotypingStrict , ChElmObservationResultsLaboratoryHivRecencyStrict , ChElmObservationResultsLaboratoryMicrobiology , ChElmObservationResultsLaboratoryMicrobiologyStrict , ChElmObservationResultsLaboratorySusceptibility , ChElmObservationResultsLaboratorySusceptibilityStrict , ChElmResultsComponentAntibioticTb , ChElmResultsComponentGeneTb , ChElmResultsLaboratoryObservation , ChElmResultsLaboratoryObservationComplete , ChElmResultsLaboratoryObservationGeno , ChElmResultsLaboratoryObservationSusc , ChElmServiceRequestLaboratoryOrder , Composition/1Comp-NeisseriaGonorrhoeae , Composition/1bComp-NeisseriaGonorrhoeae , DiagnosticReport/1DR-NeisseriaGonorrhoeae , DiagnosticReport/1bDR-NeisseriaGonorrhoeae , DiagnosticReport/1cDR-NeisseriaGonorrhoeae , DocumentReference/1-DocumentReferenceStrict , DocumentReference/1c-DocumentReferenceStrict , DocumentReference/2-DocumentReference , DocumentReference/2-DocumentReferenceStrict , DocumentReference/2-DocumentReferenceVctStrict , DocumentReference/Publish-1Doc-NeisseriaGonorrhoeae , DocumentReference/Publish-2Doc-ChlamydiaTrachomatis-Vct , DocumentReference/Publish-4Doc-Campylobacter , DocumentReference/Publish-5Doc-TreponemaPallidum , DocumentReference/Publish-6Doc-Influenza , DocumentReference/Publish-7Doc-SARSCoV2 , Observation/1Obs-NeisseriaGonorrhoeae , Observation/1bObs-NeisseriaGonorrhoeae , ServiceRequest/1SR-Order , ServiceRequest/1bSR-Order and ServiceRequest/1cSR-Order
This material contains content that is copyright of SNOMED International. Implementers of these specifications must have the appropriate SNOMED CT Affiliate license - for more information contact https://www.snomed.org/get-snomed or info@snomed.org . Show Usage
SNOMED Clinical Terms® (SNOMED CT®) : Bundle/10Doc-Legionella , Bundle/11Doc-Malaria ... Show 121 more , Bundle/12Doc-Mpox , Bundle/13Doc-Shigella , Bundle/14Doc-Neisseriameningitidis-confirmationtest , Bundle/14Doc-Neisseriameningitidis-confirmationtest-originalorder , Bundle/15Doc-Measles-Seroconversion , Bundle/16Doc-Dengue-Titer , Bundle/17Doc-Neisseria , Bundle/18Doc-C-diphtheriae , Bundle/19Doc-S-pneumoniae , Bundle/1Doc-NeisseriaGonorrhoeae , Bundle/1bDoc-NeisseriaGonorrhoeae , Bundle/1cDoc-NeisseriaGonorrhoeae , Bundle/20Doc-Vibrio-cholerae , Bundle/21Doc-HepatiteE , Bundle/22Doc-H-influenzae , Bundle/23Doc-F-tularensis , Bundle/24Doc-Chikungunya , Bundle/25Doc-Tick-borne-encephalitis , Bundle/26Doc-HepatiteA , Bundle/28Doc-Listeria-monocytogenes , Bundle/29Doc-Rubella , Bundle/2Doc-ChlamydiaTrachomatis , Bundle/2Doc-ChlamydiaTrachomatis-Vct , Bundle/30Doc-Salmonella-enteritidis , Bundle/32Doc-Rubella-avidity , Bundle/33Doc-Salmonella-valueString , Bundle/34Doc-Brucella , Bundle/35Doc-CJD , Bundle/36Doc-Salmonella-paratyphi , Bundle/37Doc-Zika , Bundle/38Doc-Anthrax , Bundle/39Doc-Botulims , Bundle/3Doc-CPE , Bundle/40Doc-Crimean-Congo , Bundle/41Doc-Ebola , Bundle/42Doc-Lassa , Bundle/43Doc-Marburg , Bundle/44Doc-Mers-CoV , Bundle/45Doc-Sars-CoV , Bundle/46Doc-Yersinia-pestis , Bundle/47Doc-Variola , Bundle/48Doc-Mpox-Clade , Bundle/49Doc-HIV , Bundle/4Doc-Campylobacter , Bundle/50Doc-HIV-viremia , Bundle/51Doc-Gelbfieber , Bundle/52Doc-Hantavirus , Bundle/53Doc-InfluenzaHxNy , Bundle/54Doc-Poliomyelitis , Bundle/55Doc-Tollwut , Bundle/56Doc-Trichinella-spiralis , Bundle/57Doc-West-Nile , Bundle/58Doc-Coxiella , Bundle/59Doc-EHEC , Bundle/5Doc-TreponemaPallidum , Bundle/61Doc-Tb-Susceptibility , Bundle/63Doc-Tb-Genotyping , Bundle/65Doc-Tuberculosis , Bundle/66Doc-HivRecency , Bundle/67Doc-Emerging-Pathogen , Bundle/6Doc-Influenza , Bundle/7Doc-SARSCoV2 , Bundle/8Doc-HepatiteB , Bundle/9Doc-HepatiteC , ChElmComposition , ChElmCompositionStrict , ChElmInterpretationCodesAvidity , ChElmInterpretationCodesSero , ChElmLabStudyTypes , ChElmObservationResultsLaboratory , ChElmObservationResultsLaboratoryHivRecencyStrict , ChElmObservationResultsLaboratoryMicrobiology , ChElmObservationResultsLaboratoryMicrobiologyStrict , ChElmResultsBruOrg , ChElmResultsCampDiarOrg , ChElmResultsCampOrg , ChElmResultsCholOrg , ChElmResultsCjdOrg , ChElmResultsCodedValuesLaboratory , ChElmResultsCompleteSpec , ChElmResultsComponentHivRecency , ChElmResultsDiphOrg , ChElmResultsGeniSpec , ChElmResultsHaemOrg , ChElmResultsHantaOrg , ChElmResultsInfluenzaHxnyOrg , ChElmResultsLaboratoryObservation , ChElmResultsLaboratoryObservationComplete , ChElmResultsLegOrg , ChElmResultsLisOrg , ChElmResultsLisSpec , ChElmResultsMalOrg , ChElmResultsMaldiTofOrg , ChElmResultsMeaOrg , ChElmResultsMenOrg , ChElmResultsMpoxOrg , ChElmResultsPneuOrg , ChElmResultsSalOrg , ChElmResultsSalOrgComplete , ChElmResultsShiOrg , ChElmResultsSterileSpec , ChElmResultsTubOrg , ChElmResultsTulOrg , ChElmResultsVirusCultOrg , ChElmServiceRequestLaboratoryOrder , ChElmSpecimen , Composition/1Comp-NeisseriaGonorrhoeae , Composition/1bComp-NeisseriaGonorrhoeae , DocumentReference/1-DocumentReferenceStrict , DocumentReference/1c-DocumentReferenceStrict , DocumentReference/2-DocumentReference , DocumentReference/2-DocumentReferenceStrict , DocumentReference/2-DocumentReferenceVctStrict , DocumentReference/Publish-1Doc-NeisseriaGonorrhoeae , DocumentReference/Publish-2Doc-ChlamydiaTrachomatis-Vct , DocumentReference/Publish-4Doc-Campylobacter , DocumentReference/Publish-5Doc-TreponemaPallidum , DocumentReference/Publish-6Doc-Influenza , DocumentReference/Publish-7Doc-SARSCoV2 , Observation/1Obs-NeisseriaGonorrhoeae and Observation/1bObs-NeisseriaGonorrhoeae
This material derives from the HL7 Terminology (THO). THO is copyright ©1989+ Health Level Seven International and is made available under the CC0 designation. For more licensing information see: https://terminology.hl7.org/license.html Show Usage
DataAbsentReason : Bundle/14Doc-Neisseriameningitidis-confirmationtest , Bundle/14Doc-Neisseriameningitidis-confirmationtest-originalorder ... Show 34 more , Bundle/15Doc-Measles-Seroconversion , Bundle/16Doc-Dengue-Titer , Bundle/18Doc-C-diphtheriae , Bundle/19Doc-S-pneumoniae , Bundle/20Doc-Vibrio-cholerae , Bundle/21Doc-HepatiteE , Bundle/22Doc-H-influenzae , Bundle/24Doc-Chikungunya , Bundle/26Doc-HepatiteA , Bundle/28Doc-Listeria-monocytogenes , Bundle/29Doc-Rubella , Bundle/32Doc-Rubella-avidity , Bundle/34Doc-Brucella , Bundle/35Doc-CJD , Bundle/37Doc-Zika , Bundle/38Doc-Anthrax , Bundle/39Doc-Botulims , Bundle/3Doc-CPE , Bundle/40Doc-Crimean-Congo , Bundle/41Doc-Ebola , Bundle/42Doc-Lassa , Bundle/43Doc-Marburg , Bundle/44Doc-Mers-CoV , Bundle/45Doc-Sars-CoV , Bundle/46Doc-Yersinia-pestis , Bundle/47Doc-Variola , Bundle/49Doc-HIV , Bundle/51Doc-Gelbfieber , Bundle/54Doc-Poliomyelitis , Bundle/55Doc-Tollwut , Bundle/56Doc-Trichinella-spiralis , Bundle/59Doc-EHEC , Bundle/63Doc-Tb-Genotyping and Bundle/67Doc-Emerging-Pathogen Observation Category Codes : Bundle/10Doc-Legionella , Bundle/11Doc-Malaria ... Show 85 more , Bundle/12Doc-Mpox , Bundle/13Doc-Shigella , Bundle/14Doc-Neisseriameningitidis-confirmationtest , Bundle/14Doc-Neisseriameningitidis-confirmationtest-originalorder , Bundle/15Doc-Measles-Seroconversion , Bundle/16Doc-Dengue-Titer , Bundle/17Doc-Neisseria , Bundle/18Doc-C-diphtheriae , Bundle/19Doc-S-pneumoniae , Bundle/1Doc-NeisseriaGonorrhoeae , Bundle/1bDoc-NeisseriaGonorrhoeae , Bundle/1cDoc-NeisseriaGonorrhoeae , Bundle/20Doc-Vibrio-cholerae , Bundle/21Doc-HepatiteE , Bundle/22Doc-H-influenzae , Bundle/23Doc-F-tularensis , Bundle/24Doc-Chikungunya , Bundle/25Doc-Tick-borne-encephalitis , Bundle/26Doc-HepatiteA , Bundle/28Doc-Listeria-monocytogenes , Bundle/29Doc-Rubella , Bundle/2Doc-ChlamydiaTrachomatis , Bundle/2Doc-ChlamydiaTrachomatis-Vct , Bundle/30Doc-Salmonella-enteritidis , Bundle/32Doc-Rubella-avidity , Bundle/33Doc-Salmonella-valueString , Bundle/34Doc-Brucella , Bundle/35Doc-CJD , Bundle/36Doc-Salmonella-paratyphi , Bundle/37Doc-Zika , Bundle/38Doc-Anthrax , Bundle/39Doc-Botulims , Bundle/3Doc-CPE , Bundle/40Doc-Crimean-Congo , Bundle/41Doc-Ebola , Bundle/42Doc-Lassa , Bundle/43Doc-Marburg , Bundle/44Doc-Mers-CoV , Bundle/45Doc-Sars-CoV , Bundle/46Doc-Yersinia-pestis , Bundle/47Doc-Variola , Bundle/48Doc-Mpox-Clade , Bundle/49Doc-HIV , Bundle/4Doc-Campylobacter , Bundle/50Doc-HIV-viremia , Bundle/51Doc-Gelbfieber , Bundle/52Doc-Hantavirus , Bundle/53Doc-InfluenzaHxNy , Bundle/54Doc-Poliomyelitis , Bundle/55Doc-Tollwut , Bundle/56Doc-Trichinella-spiralis , Bundle/57Doc-West-Nile , Bundle/58Doc-Coxiella , Bundle/59Doc-EHEC , Bundle/5Doc-TreponemaPallidum , Bundle/61Doc-Tb-Susceptibility , Bundle/63Doc-Tb-Genotyping , Bundle/65Doc-Tuberculosis , Bundle/66Doc-HivRecency , Bundle/67Doc-Emerging-Pathogen , Bundle/6Doc-Influenza , Bundle/7Doc-SARSCoV2 , Bundle/8Doc-HepatiteB , Bundle/9Doc-HepatiteC , ChElmObservationResultsLaboratory , ChElmObservationResultsLaboratoryGenotyping , ChElmObservationResultsLaboratoryGenotypingStrict , ChElmObservationResultsLaboratoryHivRecencyStrict , ChElmObservationResultsLaboratoryMicrobiology , ChElmObservationResultsLaboratoryMicrobiologyStrict , ChElmObservationResultsLaboratorySusceptibility , ChElmObservationResultsLaboratorySusceptibilityStrict , DocumentReference/1-DocumentReferenceStrict , DocumentReference/1c-DocumentReferenceStrict , DocumentReference/2-DocumentReference , DocumentReference/2-DocumentReferenceStrict , DocumentReference/2-DocumentReferenceVctStrict , DocumentReference/Publish-1Doc-NeisseriaGonorrhoeae , DocumentReference/Publish-2Doc-ChlamydiaTrachomatis-Vct , DocumentReference/Publish-4Doc-Campylobacter , DocumentReference/Publish-5Doc-TreponemaPallidum , DocumentReference/Publish-6Doc-Influenza , DocumentReference/Publish-7Doc-SARSCoV2 , Observation/1Obs-NeisseriaGonorrhoeae and Observation/1bObs-NeisseriaGonorrhoeae Test script operation code : Test100Kosovo , Test101InvalidCountryCode ... Show 8 more , Test92WarningBerUidGln , Test93ErrorFullNameInfluenca , Test94IgnoreSourceWarning , Test95IgnoreObservationInterpretationCodesWarning , Test96IgnoreObservationInterpretationCodesInformation , Test97CoaxiellaNoRatio , Test98HivInvalidCode and Test99CoaxiellaZeroDenominator identifierType : ChElmPatient , ChElmPatientHIV , ChElmPatientInitials and ChElmPatientVCT ObservationInterpretation : Bundle/10Doc-Legionella , Bundle/11Doc-Malaria ... Show 77 more , Bundle/12Doc-Mpox , Bundle/13Doc-Shigella , Bundle/14Doc-Neisseriameningitidis-confirmationtest , Bundle/14Doc-Neisseriameningitidis-confirmationtest-originalorder , Bundle/16Doc-Dengue-Titer , Bundle/17Doc-Neisseria , Bundle/18Doc-C-diphtheriae , Bundle/19Doc-S-pneumoniae , Bundle/1Doc-NeisseriaGonorrhoeae , Bundle/1bDoc-NeisseriaGonorrhoeae , Bundle/1cDoc-NeisseriaGonorrhoeae , Bundle/20Doc-Vibrio-cholerae , Bundle/21Doc-HepatiteE , Bundle/22Doc-H-influenzae , Bundle/23Doc-F-tularensis , Bundle/24Doc-Chikungunya , Bundle/25Doc-Tick-borne-encephalitis , Bundle/26Doc-HepatiteA , Bundle/28Doc-Listeria-monocytogenes , Bundle/2Doc-ChlamydiaTrachomatis , Bundle/2Doc-ChlamydiaTrachomatis-Vct , Bundle/30Doc-Salmonella-enteritidis , Bundle/33Doc-Salmonella-valueString , Bundle/34Doc-Brucella , Bundle/35Doc-CJD , Bundle/36Doc-Salmonella-paratyphi , Bundle/37Doc-Zika , Bundle/38Doc-Anthrax , Bundle/39Doc-Botulims , Bundle/3Doc-CPE , Bundle/40Doc-Crimean-Congo , Bundle/41Doc-Ebola , Bundle/42Doc-Lassa , Bundle/43Doc-Marburg , Bundle/44Doc-Mers-CoV , Bundle/45Doc-Sars-CoV , Bundle/46Doc-Yersinia-pestis , Bundle/47Doc-Variola , Bundle/48Doc-Mpox-Clade , Bundle/49Doc-HIV , Bundle/4Doc-Campylobacter , Bundle/50Doc-HIV-viremia , Bundle/51Doc-Gelbfieber , Bundle/52Doc-Hantavirus , Bundle/53Doc-InfluenzaHxNy , Bundle/54Doc-Poliomyelitis , Bundle/55Doc-Tollwut , Bundle/56Doc-Trichinella-spiralis , Bundle/57Doc-West-Nile , Bundle/58Doc-Coxiella , Bundle/59Doc-EHEC , Bundle/5Doc-TreponemaPallidum , Bundle/61Doc-Tb-Susceptibility , Bundle/63Doc-Tb-Genotyping , Bundle/65Doc-Tuberculosis , Bundle/67Doc-Emerging-Pathogen , Bundle/6Doc-Influenza , Bundle/7Doc-SARSCoV2 , Bundle/8Doc-HepatiteB , Bundle/9Doc-HepatiteC , ChElmInterpretationCodesPos , ChElmInterpretationCodesPosNeg , ChElmInterpretationCodesResSus , ChElmInterpretationCodesTiter , DocumentReference/1-DocumentReferenceStrict , DocumentReference/1c-DocumentReferenceStrict , DocumentReference/2-DocumentReference , DocumentReference/2-DocumentReferenceStrict , DocumentReference/2-DocumentReferenceVctStrict , DocumentReference/Publish-1Doc-NeisseriaGonorrhoeae , DocumentReference/Publish-2Doc-ChlamydiaTrachomatis-Vct , DocumentReference/Publish-4Doc-Campylobacter , DocumentReference/Publish-5Doc-TreponemaPallidum , DocumentReference/Publish-6Doc-Influenza , DocumentReference/Publish-7Doc-SARSCoV2 , Observation/1Obs-NeisseriaGonorrhoeae and Observation/1bObs-NeisseriaGonorrhoeae RoleCode : ChElmPatient , ChElmPatientHIV , ChElmPatientInitials and ChElmPatientVCT
Cross Version Analysis
This is an R4 IG. None of the features it uses are changed in R4B, so it can be used as is with R4B systems. Packages for both R4 (ch.fhir.ig.ch-elm.r4) and R4B (ch.fhir.ig.ch-elm.r4b) are available.
Dependencies
Dependency Overview
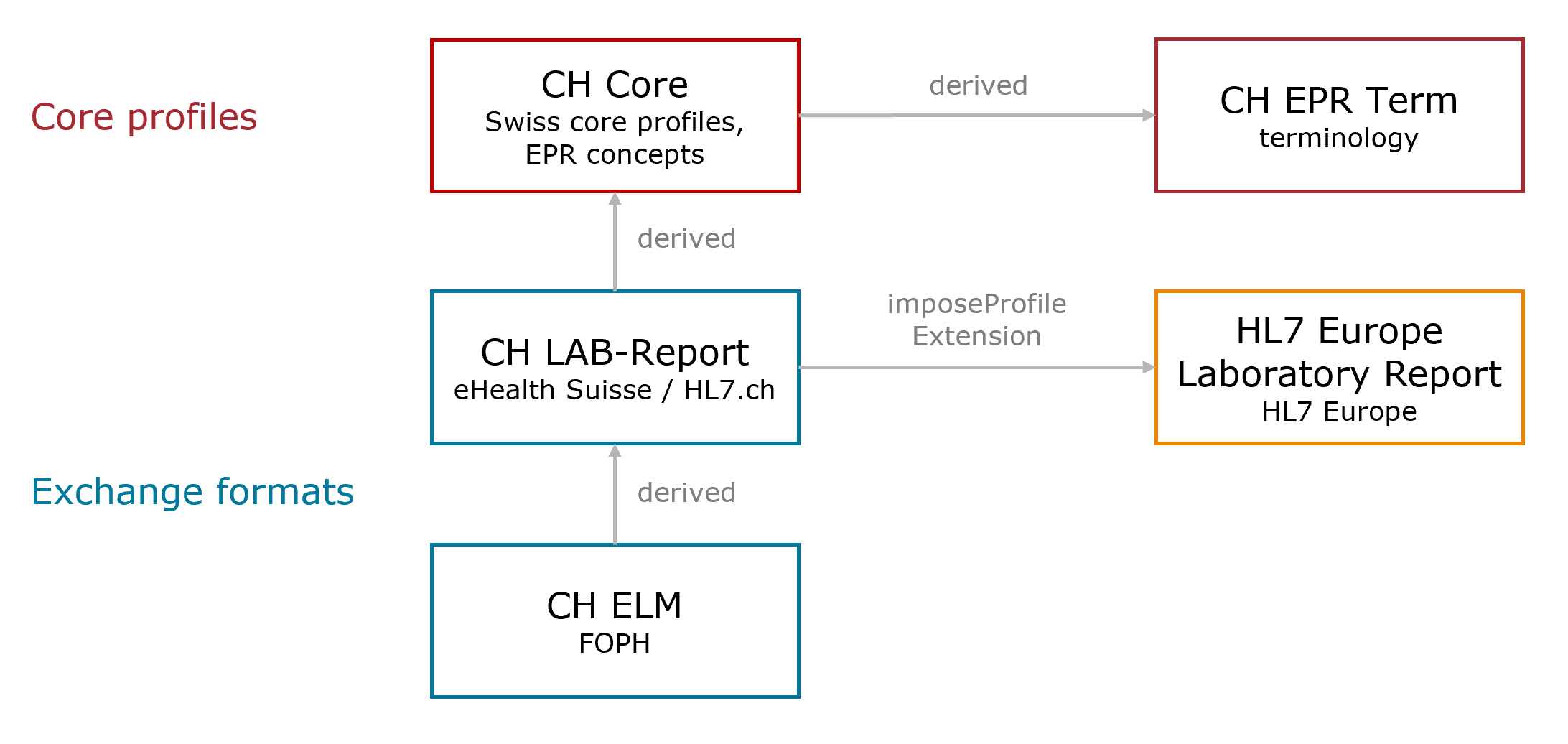
This overview illustrates the relevant dependencies of CH ELM to the Swiss implementation guides and the European laboratory project .
Fig. 1: Dependency Overview
Dependency Table
Package hl7.fhir.uv.extensions.r4#5.2.0
This IG defines the global extensions - the ones defined for everyone. These extensions are always in scope wherever FHIR is being used (built Mon, Feb 10, 2025 21:45+1100+11:00)
Package hl7.fhir.uv.ips#1.1.0
International Patient Summary (IPS) FHIR Implementation Guide (built Tue, Nov 22, 2022 03:24+0000+00:00)
Package hl7.fhir.eu.laboratory#0.1.1
This guide describes how the Laboratory Report can be represented in the European REALM. (built Tue, Mar 25, 2025 12:00+0100+01:00)
Package ch.fhir.ig.ch-core#6.0.0-ballot
FHIR implementation guide CH Core (built Thu, May 22, 2025 05:56+0000+00:00)
Package hl7.fhir.uv.sdc#3.0.0
The SDC specification provides an infrastructure to standardize the capture and expanded use of patient-level data collected within an EHR.
Package ch.fhir.ig.ch-orf#3.0.1
Order & Referral by Form - Implementation Guide (CH ORF) (built Thu, May 22, 2025 07:53+0000+00:00)
Package hl7.fhir.uv.order-catalog#0.1.0
An Order Catalog is an administered homogeneous collection of items such as medication products, laboratory tests, procedures, medical devices or knowledge artifacts such as order sets, which support the ordering process, or more generally healthcare processes. (built Sun, Aug 9, 2020 15:18+0000+00:00)
Package hl7.fhir.uv.extensions.r4#1.0.0
This IG defines the global extensions - the ones defined for everyone. These extensions are always in scope wherever FHIR is being used (built Sun, Mar 26, 2023 08:46+1100+11:00)
Package hl7.fhir.eu.extensions#0.1.0
This guide lists the extensions speciifed for the European REALM. (built Tue, Feb 20, 2024 08:56+0100+01:00)
Package ch.fhir.ig.ch-lab-report#2.0.0-ballot
FHIR® Implementation Guide for Laboratory Reports in Switzerland (built Thu, May 22, 2025 14:08+0000+00:00)
Package hl7.fhir.uv.tools.r4#0.9.0
This IG defines the extensions that the tools use internally. Some of these extensions are content that are being evaluated for elevation into the main spec, and others are tooling concerns (built Tue, Dec 16, 2025 23:18+1100+11:00)
Globals Table
There are no Global profiles defined
![]() ® are trademarks owned by Health Level Seven International, registered with the United States Patent and Trademark Office.
® are trademarks owned by Health Level Seven International, registered with the United States Patent and Trademark Office.